IC 之音專訪(清大的產學合作及智慧生醫):https://www.facebook.com/story.php?story_fbid=pfbid0Wb8Y94h2DQpGtCCPQDuzz1y59bYAEKFDAFkGnPaEEkkJiLF1EJWffVarv2Z8TZEql&id=100063884146466&mibextid=qC1gEa&paipv=0&eav=AfZhp8JzUL8cmOWzj5gLCKZZTHsOvJFAXOBXyQzRHIeRld0XctGrCs21q_i0qExOoZU&_rdr (Dec 2023 & Mar 2024)
Our new sensor study co-first-authored and co-corresponding with Prof Lin, Zhong-Hong’s group just got accepted in Nano Energy 🙏 Congratulations to Anny, Pramod and Christopher – Yu-Ying Cheng#, Anindita Ganguly#, Yi-Yun Cheng#, Christopher Llynard D. Ortiz, Arnab Pala, Pramod Shah, Lee-Wei Yang*, Zong-Hong Lin* (2024) Development of Label-Free Triboelectric Nanosensors as Screening Platforms for Anti-Tumor Drugs. Nano Energy (accepted) (5-yr IF: 17.5; Ranking: 11/161 in Chemistry, Physical) (Mar 2024)
European Patent “PRIVACY-KEPT TEXT COMPARISON METHOD, SYSTEM AND COMPUTER PROGRAM PRODUCT” was granted. 歐洲發明專利申請案-核准通知:V9P108002-EU/2012-07-425(清大)19(專)A065-EP(交大)_ P198914EPI_ 申請人:國立清華大學、國立交通大學 發 明 人:楊立威、洪瑞鴻、艾曼紐、張芫瑜 案件名稱:文字比對方法、系統及其電腦程式產品 申 請 日:2020/06/25 申 請 號:20 182 227.7 (Mar 2024)
Congratulations to our former lab members Cheng-Yu Tsai and Yuan-Yu Chang for having a mechanistic study on pathogenicity of a popular gene variant in hearing impairment accepted by Computational and Structural Biotechnology Journal – Cheng-Yu Tsai, Ying-Chang Lu, Yen-Hui Chan, Navaneethan Radhakrishnan, Yuan-Yu Chang, Shu-Wha Lin, Tien-Chen Liu, Chuan-Jen Hsu, Pei-Lung Chen, Lee-Wei Yang*, Chen-Chi Wu* (2023) Simulation-Predicted and -Explained Inheritance Model of Pathogenicity Confirmed by Transgenic Mice Models. Computational and Structural Biotechnology Journal https://www.sciencedirect.com/science/article/pii/S2001037023004415 (5-yr IF: 6.3) (Nov 2023)
Congratulations to PhD students Ahmed Ragab and Chris L Ortiz who won the 1st Best and 2nd Best Oral Presentation Award respectively, in the annual institutional retreat (Inst. Bioinformatics and Structural Biology) Master student Mr Chih-Yuan Kao won the Best Poster Award (Sept 2023). 


 歡迎報考清大智慧生醫博士學位學程 (Welcome to apply for BAI PhD program, NTHU):
歡迎報考清大智慧生醫博士學位學程 (Welcome to apply for BAI PhD program, NTHU):
https://news.gbimonthly.com/tw/magazine/article_show.php?num=60698&page=1&tag=%E5%B0%81%E9%9D%A2%E6%95%85%E4%BA%8B
https://bai.site.nthu.edu.tw/ (Aug 2023)
After efforts more than a decade. the first Taiwanese-made full-text search engine, S.A.V.E. for plagiarism detection with privacy protection was accepted and published in IEEE Journal of Biomedical and Health Informatics (https://ieeexplore.ieee.org/document/10198547). 此具有隱私模式搜尋功能的全文比對引擎Sapiens Aperio Veritas Engine (S.A.V.E.)先將敏感文件中的文字做破壞性的編
引擎入口:
https://dyn.life.nthu.edu.tw/SAVE/
https://save.praexisio.com.tw (2020年技轉)
文章連結:https://ieeexplore.ieee.org/document/10198547
台美專利:
https://patents.google.com/patent/US11232157B2/en (US11232157B2)
https://patents.google.com/patent/TWI719537B/en(TWI719537B)
(July 2023)
Congratulations to Mr Christopher L Ortiz to win the poster presentation competition in 2023 international conference, “Nucleic Acids: Prospect and Therapeutic Applications”, held in National Chung Hsin University. The presentation topic is “Frameshift-promoting PK induces a Rolled and Hyper-rotated 70S Ribosome”. 恭喜博班Chris Ortiz同學榮獲2023核酸結構研究前景與醫學應用國際研討會海報比賽優勝 (July 2023) 
Congratulations to Mr Ian Kao to win the poster competition in 2023 Graduate Student Symposium in the College of Life Science and Medicine, NTHU;恭喜碩班高祺原同學榮獲2023清大生命科學院研究生學術研討會海報比賽優勝  (May 2023)
(May 2023)
One article on interplay between protein dynamics and machine learning, co-authored with Prof Ivet Bahar, was recently accepted by Current Opinion in Structural Biology, entitled “Mutually Beneficial Confluence of Structure-Based Modeling of Protein Dynamics and Machine Learning Methods” by Anupam Banerjee, Satyaki Saha, Nathan C. Tvedt, Lee-Wei Yang and Ivet Bahar. https://doi.org/10.1016/j.sbi.2022.102517 (Nov 2022)
Congratulations to our previous RA Mr Lin, Hong-Rui for having a co-authored paper on centrosome diffusion barrier accepted by EMBO Report. “Actin filaments form a size-dependent diffusion barrier around centrosomes” by Hsuan Cheng, Yu-Lin Kao, Ting Chen, Lohitaksh Sharma, Wen-Ting Yang, Yi-Chien Chuang, Shih-Han Huang, Hong-Rui Lin, Yao-Shen Huang, Ching-Lin Kao, Lee-Wei Yang, Rachel Bearon, Hui-Chun Cheng, Kuo-Chiang Hsia, and Yu-Chun Lin. EMBO Report 24, e54935, where the leading PI is Prof. Lin, Yu-Chun (Oct 2022)
Congratulations to our undergrade student Mr 曾鴻明 to win the 2022 Undergrad Research Scholarship provided by CLS, NTHU (111學年度清華大學生命科學院大學生進行專題研究計畫獎學金獲補助) and obtain 30,000 NTD scholarship for his research on Druggable DynOmics – “可藥動態組的推動:30個可溶蛋白和5個膜蛋白構型探索與分析以及小分子藥物力場正確性的確認” (Aug 2022)
Congratulations to our Master-degree student Ms Yun-Pei Chang (張芸霈) to win the Best Oral Presentation Award and ~2600 NTD cash prize in the Annual Retreat of the Institute of Bioinformatics and Structural Biology 2022 (Jun 2022)
Domestic news related to the functional peptide design (Mar 2022) –
1.清華首頁:https://www.nthu.edu.tw
2.清華FB:https://www.facebook.co
3.清華IG:https://www.instagram.c
4.聯合報:https://udn.com/news/sto
5.自由時報:https://health.ltn.com.
6.中央社:https://www.cna.com.tw/n
7.中時新聞:https://www.chinatimes.
8.科技新報:https://technews.tw/202
9.中華新聞:https://www.cdns.com.tw
10.新浪新聞:https://news.sina.com.
US patent granted – PRIVACY-KEPT TEXT COMPARISON METHOD, SYSTEM AND COMPUTER PROGRAM PRODUCT/ 隱私保護全文搜尋引擎美國專利獲證 https://patentscope.wipo.int/search/en/detail.jsf?docId=US315292148 (Jan 2022) (the preprint that summarizes our technology – https://arxiv.org/abs/2201.00696 )
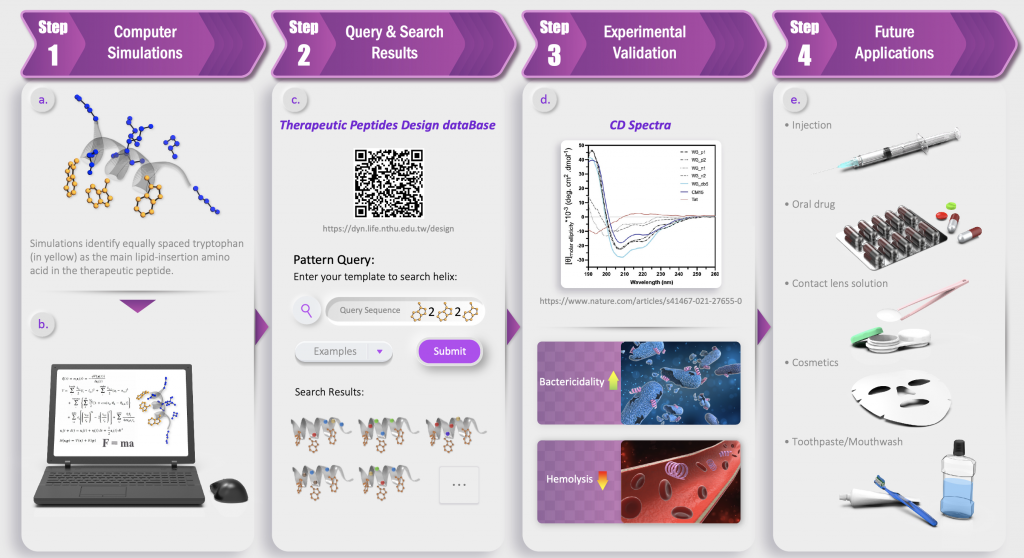
Congratulations to our former and current members Cheng-Yu, Liyin, Kai-Di, Sohba, Lutimba, Chris, Yu-Lin, summer intern students: Adarsh, Ximai as well as Drs Emmanuel Salawu, Hongchun Li, Yuan-Yu Chang and Anny Cheng for reporting their therapeutic peptide design story at Nature Communications – Cheng-Yu Tsai, Emmanuel O Salawu, Hongchun Li, Guan-Yu Lin, Ting-Yu Kuo, Liyin Voon, Adarsh Sharma, Kai-Di Hu, Yi-Yun Cheng, Sobha Sahoo, Lutimba Stuart, Chih-Wei Chen, Yuan-Yu Chang, Yu-Lin Lu, Ximai Ke, Christopher Llynard D. Ortiz, Bai-Shan Fang, Chen-Chi Wu, Chung-Yu Lan*, Hua-Wen Fu*, Lee-Wei Yang* (2022) Helical Structure Motifs Made Searchable to Facilitate the Functional Peptide Design. Nature Communications, 13, Article number: 102. https://www.nature.com/articles/s41467-021-27655-0 (Dec 2021)
Congratulations to our former and current members Hongrui Lin and Christopher Llynard Ortiz published their antinociceptive peptide design in eLife together with the UT Southwestern team – Wentong Fang, Chengheng Liao, Giada Zurlo, Jeremy Simon, Travis Ptacek, Youqiong Ye, Leng Han, Cheng Fan, Christopher Ortiz, Hongrui Lin, Ujjawal Manocha, William Kim, and Lee-Wei Yang and Qing Zhang (2021) ZHX2 Promotes HIF1a Oncogenic Signaling in Triple-Negative Breast Cancer. eLife https://elifesciences.org/articles/70412 (Nov 2021)
Congratulations to our former and current members Yi-Hsuan Tsai, Yuan-Yu Chang and Christopher Llynard Ortiz published their antinociceptive peptide design in Pharmaceutics together with the UBC team – K K DurgaRao Viswanadham, Roland Böttger, Lukas Hohenwarter, Anne Nguyen, Elham Rouhollahi, Alexander Smith, Yi-Hsuan Tsai, Yuan-Yu Chang, Christopher Llynard Ortiz, Lee-Wei Yang, Liliana Jimenez, Siyuan Li, Chan Hur, Shyh-Dar Li (2021) An Effective and Safe Enkephalin Analog for Antinociception. Pharmaceutics, 13, 927- (June 2021)
3000 citations achieved from 35 SCI journal papers and 3 non-SCI papers (Sept 23, 2020)
Our lab obtained the US patent of “(US20190183860) METHOD FOR THE TREATMENT OF ATG4-RELATED DISORDERS”
|
||||
|
| Applicants: | National Tsing Hua University |
| Inventors: | Lee-Wei Yang Chih-Wen Shu Pei-Feng Liu Kun-Lin Tsai |
本實驗室獲得ATG4相關抗癌藥物之美國專利! (Dec 2019)
Our New Publication on Linear Response Theory (https://www.jstage.jst.go.jp/article/biophysico/16/0/16_473/_article/-char/en ); the work is also in honor of the contribution of Prof Nobuhiro Go to the Progress of Theoretical and Computational Biophysics (Nov 2019)
Congratulations to our another linear response theory paper getting accepted – Bang-Chieh Huang (our former postdoc) and Lee-Wei Yang (2019) Molecular dynamics simulations and linear response theories jointly describe biphasic responses of myoglobin relaxation and reveal evolutionarily conserved frequent communicators. (accepted by Biophysics and Physicobiology; a special issue to honor Nobuhiro Gō sensei’s contribution to Biophysics Society and his 80 year old birthday) bioRxiv (Sept 2019)
Congratulations to Dr Justin Chan for his latest first-author publication accepted by Structure – Justin Chan, Hong-Rui Lin, Kazuhiro Takemura, Kai-Chun Chang, Yuan-Yu Chang, Yasumasa Joti, Akio Kitao, Lee-Wei Yang* (2019) “An efficient timer and sizer of biomacromolecular motions”. Structure (accepted; preprint) (Sept 2019)
Congratulations to Mr Kevin Chen (陳柏淳) for passing the oral defense for his Master degree and thesis entitled “A novel software to predict the -1 Programmed Ribosomal Frameshifting (-1PRF) signals in viral genomes –predicted signals are validated by cell-free bioassays for Zika virus genome” ! (July 2019)
Congratulations to Dr-to-be Justin Chan for winning the best student oral presentation award in 2019 Computational Biology Workshop held by National Center for Theoretical Sciences (NCTS) as well as his first-author publication accepted by Bioinformatics – Justin Chan, Jinhao Zou, Christopher Llynard Ortiz, Chi-Hong Chang Chien, Rong-Long Pan, Lee-Wei Yang* (2019) DR-SIP: Protocols for Higher Order Structure Modeling with Distance Restraints- and Cyclic Symmetry-Imposed Packing. Bioinformatics (accepted) https://doi.org/10.1093/bioinformatics/btz579 (July 2019)
Congratulations to Mr Kun-Wei Lin (林昆衛) in receiving the Merit Award in 2019 Graduate Student Symposium in the College of Life Science, NTHU (Cash award/優勝獎金:5,000 NTD) (May 2019)
Our China patent for “Evaluation system for the efficacy of antimicrobial peptides and the use thereof” has been approved.
本實驗室獲得中國專利核准通過 CN106469251A
校內編號:V9P104070-CN
本所案號:THPA-2014004
國別/類別:中國 / 發明專利
專利名稱:评估抗菌肽抗菌力的系统及其使用方法
申請日期:2016.08.17
申請案號:201610682437.7
代 理 人:北京元本知識產權代理事務所
另一分案專利也已公開 CN109535226A (Apr 2019)
Congratulations to Yang lab for being awarded 48,000 node-hours computation resources by High Performance Computing Infrastructure (HPCI), Japan, for the year 2019-2020! (Feb 2019)
Congratulations to Mr Kevin Chen (陳柏淳) in receiving scholarship for entering the accelerated Master program (預研生入學獎學金:每名10,000元及獎狀乙紙) ! (Sept 2018)
Congratulations to Dr Kai-Chun Chang, Mr Emmanuel O Salawu (co-first author) and Mr Yuan-Yu Chang for their latest publication in Bioinformatics – Kai-Chun Chang* , Emmanuel Oluwatobi Salawu, Yuan-Yu Chang, Jin-Der Wen and Lee-Wei Yang* (2018) Ribosomal frameshifting is a consequence of force-induced subunit rolling. Bioinformatics. (published; IF: 7.3; ranked 3/59 in Mathematical & Computational Biology) (Aug 2018)
Congratulations to Mr Kun-Wei Lin (林昆衛) who was awarded the undergraduate research fund provided by Life Science College, NTHU (30,000 NTD a year); 恭喜 Mr 林昆衛 之研究專題「結合模擬與實驗以跨分子異構調控的方式來下調細胞自噬蛋白之活性進而抑制癌細胞的生長」獲得107學年清華大學生命科學院獎勵大學生進行專題研究計畫獲補助 (Aug 2018)
Congratulations to Mr Kun-Lin Tsai for winning the Scholarship (20,000 NTD) from Shen’s Culture & Education Foundation (沈巨塵先生清華獎學金-論文發表獎勵金) for his recent publication on Theranostics ( Dec, 2017) !
Congratulations to Ms Jinhao Zou who had her first paper accepted – Imran Khan, Yu-Kai Su, Jinhao Zou, Lee-Wei Yang, Ruey-Hwang Chou* and Chin Yu* (2017) S100B as an antagonist to block the interaction between S100A1 and the RAGE V domain. PLOS ONE, 13(2):e0190545 (accepted Dec, 2017; published Feb, 2018)
Congratulations to Mr Kun-Lin Tsai (co-first author) for his second publication from this lab – Pei-Feng Liu†; Kun-Lin Tsai†; Chien-Jen Hsu; Wei-Lun Tsai; Jin-Shiung Cheng; Hsueh-Wei Chang; Chung-Wai Shiau; Yih-Gang Goan; Ho-Hsing Tseng; Chih-Hsuan Wu; John C. Reed; Lee-Wei Yang*; Chih-Wen Shu*(2017) Drug Repurposing Screening Identifies Tc as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy, Theranostics 8, 830-845 (IF = 8.8; rank 8/128 in Medicine, Research & Experimental)(Dec, 2017) http://www.thno.org/v08p0830.htm
Congratulations to Mr Yuan-Yu Chang who just won the Scholarship (20,000 NTD) from Shen’s Culture & Education Foundation (沈巨塵先生清華獎學金-論文發表獎勵金) for his publication – “DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Research (2017)” (June, 2017)
Congratulations to Ms Jinhao (Jane) Zou who won the honorable mention (佳作) and cash prize 2000 NTD in 2017 College of Life Science Graduate Student Symposium, NTHU (June 2nd, 2017)
Congratulations to Mr Justin Chan won the second place in the poster/oral presentation competition in the 22nd Biophysics Conference, Taiwan. Mr Yuan-Yu Chang were selected to be one of the best poster presenters. (May 2017) http://biophys.sinica.edu.tw/Symposium/22BC/abstract.php (Group A)
Congratulations to Mr Yuan-Yu Chang (co-first author) for his fourth publications from this lab – Hongchun Li†, Yuan-Yu Chang†, Ji Young Lee, Ivet Bahar* and Lee-Wei Yang* (2017). DynOmics: Dynamics of Structural Proteome and Beyond. Nucleic Acids Research. 45, W374–W380 (IF: 9.2; Rank 18/289 in Biochemistry & Molecular Biology)(Apr 2017) https://academic.oup.com/nar/article/45/W1/W374/3791214?searchresult=1 ; Listed as one example of the graphical abstracts by NAR – https://academic.oup.com/nar/pages/graphical_abstracts_examples & being the first paper mentioned in its category – https://academic.oup.com/nar/article/45/W1/W1/3896331
US & China patent publication (https://www.google.com/patents/US20170061288 https://www.google.com/patents/CN106469251A) (Mar 2017)
Congratulations to Mr Chris Lo (co-first author) and Mr Yuan-Yu Chang for their first and third publications from this lab – Jui-Chieh Yin, Chun-Hsien Fei, Yen-Chen Lo, Yu-Yuan Hsiao, Jyun-Cyuan Chang, Jay C Nix, Yuan-Yu Chang, Lee-Wei Yang*, I-Hsiu Huang*, Shuying Wang* (2016, Nov) Structural insights into substrate recognition by Clostridium difficile sortase. Frontiers in Cellular and Infection Microbiology. (IF: 5.2; Rank 18/123 in Microbiology)(Nov 2016)
We obtained the first preliminary approval of our China patent (中國 / 發明專利專利名稱:评估抗菌肽抗菌力的系统及其使用方法申請日期:2016.08.17 申請案號:201610682437.7 申請人:楊立威 代理人:北京元本知識產權代理事務所 審查人員:李煒倩) (Oct 2016)
Congratulations to Mr Kun-Lin Tsai in obtaining Chancellor’s Scholarship (校長獎學金; 25,000NT/month) for 4 years to support his PhD study! (Oct 2016)
Congratulations to Aravind and Justin’s recent publication in Journal of Chemical Theory and Computation – Chandrasekaran, Aravind, Chan, Justin, Lim, Carmay*, Yang, Lee-Wei* (2016) Protein dynamics and contact topology reveal protein-DNA binding orientation. Journal of Chemical Theory and Computation doi:10.1021/acs.jctc.6b00688 (5-year impact factor: 5.8; Ranking: 5/35 in Atomic, Molecular & Chemical Physics) http://pubs.acs.org/doi/full/10.1021/acs.jctc.6b00688 (Sept 2016).
We obtained the first Taiwan patent on computational evaluation system for the efficacy of antimicrobial peptide (Sept 2016)!
證書號/公告號 I557587 公開號 201709097 國別/類別:中華民國 / 發明專利 專利名稱:評估抗菌胜肽之抗菌力之系統及其使用方法 申請日期:2015.08.17 申請案號:104126710 申 請 人:國立清華大學 發 明 人:楊立威、程家維、李紅春、蔡政育、游輝元 代 理 人:王正利 審查人員:謝進忠
領證期限:2016.12.06
Congratulations to Mr Kevin Chen (陳柏淳) in receiving College Research Support in 2016 (105學年度清華大學生命科學院獎勵大學生進行專題研究計畫獲補助) for the ZIKA virus related research, entitled “以生物資訊、電腦模擬、單分子光鉗以及cell-free系統來偵測Zika病毒的框架移轉(ribosomal frameshift)位置,偽節(pseudoknot)的二級結構及其機械抗力”! (Sept 2016)
Congratulations to Mr Kun-Lin Tsai’s recent publication in BBA – Chin-Chi Chang; Md Imran Khan; Kun-Lin Tsai; Hongchun Li; Lee-Wei Yang*; Ruey-Hwang Chou*; Chin Yu* (2016) Blocking the interaction between S100A9 and RAGE V domain using CHAPS molecule: A novel route to drug development against cell proliferation. BBA – Proteins and Proteomics [impact factor = 3.3]! (Aug 2016)
Congratulations to Mr Yuan-Yu Chang who won the honorable mention (佳作) in oral presentation of published papers at 2016 NTHU&NCTU graduate students’ academic conference (2016 清交生科院研究生學術研討會) (May 2016) !
Congratulations to Mr Emmanuel O Salawu’s and Mr Yuan-Yu Chang’s co-authored Scientific Reports paper got accepted! Wen-Ting Chen, Wen-Yang Huang, Ting Wen, Emmanuel Oluwatobi Salawu, Dongli Wang, Yi-Zong Lee, Yuan-Yu Chang, Lee-Wei Yang, Shih-Che Sue, Xinquan Wang, and Hsien-Sheng Yin* (2016) Structure and function of chicken interleukin-1 beta mutants: uncoupling of receptor binding and in vivo biological activity. Scientific Reports [impact factor = 5.3] (May 2016) !
Congratulations to Mr Kai-Chun Chang who won the first prize of the poster competition in the 21st Biophysics Society Meeting in Taiwan (May 2016) !
Congratulations to Hongchun Li and Yuan-Yu Chang for the acceptance of their recent work published in NAR – Hongchun Li, Yuan-Yu Chang, Yang, L-W.* and Ivet Bahar* (2015) iGNM 2.0: The Gaussian Network Model Database for Biomolecular Structural Dynamics. Nucleic Acids Research, doi: 10.1093/nar/gkv1236 (Dec 2015) [impact factor = 9.1] !! (Dec 2015)
Congratulations to Mr Chris Yen-Chen Lo who was chosen and given scholarship to attend the Higher European Research Course for Users of Large Experimental Systems (HERCULES) 2016 Course from Tuesday March 29th to Friday April 29th, on session B including 6 days in Paris at SOLEIL Synchrotron or Laboratoire Léon Brillouin or in Switzerland at Swiss Light Source or in Hamburg at DESY or in Italy at Elettra in Trieste (Dec 2015) !!
Congratulations to Mr Yuan-Yu Chang became one of the three 2015 LS college Ph.D new student scholarship awardees (104學年度生科院博士班新生獎學金; 24,000NT/month) !!! (Oct 2015)
2015兩岸四地大學排名
A new Review article from Kai-Chun Chang, Profs Wen and Yang was just published Kai-Chun Chang, Jin-Der Wen and Lee-Wei Yang (2015) Functional Importance of Mobile Ribosomal Proteins. BioMed Research International, V2015, Article ID 539238
http://www.hindawi.com/journals/bmri/aa/539238/ (Sept 2015)
CONGRATULATIONS to Mr 張簡啟宏 who successfully defended his Master thesis. (July 2015) !!
Congratulations to Mr Yuan-Yu Chang for obatining a poster award (among Master student participants) in the 2015 College of Life Science Graduate Student Sympoisum, NTHU (2000NT cash prize)(June 2015)
Congratulations to Mr Mick Lin for obatining the undergraduate research fund provided by Ministry of Science and Technology (48K NT for a year); 恭喜 Mr 林泓叡之研究專題「發展以結構和動態為主的物理指標來預測分子演化的保守性」獲得104學年科技部獎勵大專生進行專題研究計畫獲補助 (May 2015)
CONGRATULATIONS to Mr 張芫瑜 who successfully defended his Master thesis and passed the PhD entrance exam of NTHU (結構生物學程). (May 2015)
Congratulations to Dr Bang-Chiech Huang obatining Poster Award in 第六屆海峽兩岸理論與計算化學(Theoretical and Computational Chemistry)會議壁報論文展 for the title “Intramolecular communications and allosteric regulations revealed by linear response theories” (Jan 2015)
Congratulations to Dr Bang-Chiech Huang obatining MOST postdoc fellowship for a year (~1M a year); 恭喜 黃邦杰博士獲得「延攬科技人才及兩岸科技交流(延攬博士後研究人才)」補助一年 (Nov 2014)
Congratulations to Mr Kun-Lin Tsai obatining Scholarship of Shen’s Culture & Education Foundation (50K for 10 months); 恭喜 Mr 蔡坤霖獲得103學年度沈巨塵先生清華獎學金「優秀學生研究獎學金」 (Nov 2014)
Congratulations to Mr Mick Lin obatining the undergraduate research fund provided by Life Science College (30K for a year); 恭喜 Mr 林泓叡 之研究專題「藉由蛋白結構中原子間作用力觀察與演化上的比對建立結構與演化之間的關係」獲得103學年清華大學生命科學院獎勵大學生進行專題研究計畫獲補助; 獲獎人中唯一十六級的學生(其餘為十五級) (Sept 2014)
CONGRATULATIONS to Dr Hongchun Li who just defended the thesis and obatined his PhD degree as well as accepted an offer from University of Pittsburgh, as a postdoctoral fellow, to continue his work on protein conformational dynamics and drug design. [[[狂賀!!!]]] 博班生李紅春獲得博士學位(28歲)並即將在十一月底前往匹茲堡大學醫學院進行博士後研究。(Sept 2014)
Ms Jia-Chen Wu accepted the offer of a RA position to study the energy basis for enantiomer selectivity and develop a theoretical platform to predict relevant enzymes’ specificity/selectivity for different substrates in Prof S.W. Tsai’s lab (Aug 2014)
New member was born !!! Dr Ban-Chieh Huang’s third boy was born at 5:00 pm, 7/24, 2014. Congratulations !!! (July 2014)
New articles accepted (July 2014)
Hongchun Li, Shun Sakuraba, Aravind Chandrasekaran and Lee-Wei Yang* (2014) Molecular binding sites are located near the interface of intrinsic dynamics domains (IDDs) J.Chem. Inf. Model. (IF: 4.3; Ranking: 3/100 in Computer Science)(accepted) http://pubs.acs.org/doi/abs/10.1021/ci500261z
Lee W Yang, Akio Kitao, Bang-Chieh Huang and Nobuhiro Gō (2014) Ligand-induced Protein Responses and Mechanical Signal Propagation described by Linear Response Theories. Biophysical Journal, 107 (IF: 4.0)(accepted) http://www.sciencedirect.com/science/article/pii/S0006349514007954#
Mr Aravind Chandrasekaran decided to accept the offer provided by Dept of Chemistry in U Maryland to continue his academic pursue, as a PhD student, in Computational Biology/Theretical Chemistry (Jun 2014)
Mr Aravind Chandrasekaran obtained admissions of PhD programs in University of Michigan at Ann Arbor, Baylor college of Medicine, University of Maryland and CMU-Pitt to continue his academic endeavors in Computational Biology/Chemistry (Mar 2014)
Ms Jia-Chen Wu successfully defended her Master thesis for her good work in the applicability of time-independent linear response theory for protein-ligand binding (Feb 2014)
Mr Aravind Chandrasekaran successfully defended his Master thesis for his good work on DNA – Protein interactions as a function of protein dynamics and contact topology (Feb 2014)
Mr Yen-Hua Chang successfully defended his Master thesis for his good work on predictions of enzyme active sites using partial least square regression over a set of sequence, structure and dynamics features (Oct 2013)
New article accepted (Sept 2013)
Arun A. Gupta, Ruey-Hwang Chou, Hongchun Li, Lee-Wei Yang, Chin Yu, Structural Insights into the Interaction of Human S100B and Basic Fibroblast Growth Factor (FGF2): Effects on FGFR1 Receptor Signaling, BBA – Proteins and Proteomics (2013), doi: 10.1016/j.bbapap.2013.09.012
New article published (May 2013)
Title: The N-terminal substrate-recognition domain of a LonC protease exhibits structural and functional similarity to cytosolic chaperones
Author(s): Jhen-Kai Li, Jiahn-Haur Liao, Hongchun Li, Chiao-I Kuo, Kai-Fa Huang, Lee-Wei Yang, Shih-Hsiung Wu and Chung-I Chang*has been accepted for publication in Acta Crystallographica Section D (2013). D69, 1789–1797
New Editorial published (Dec 2012)
Lee-Wei Yang, Silvina Matysiak, Shang-Te Danny Hsu, Gabriela Mustata Wilson, and Yasumasa Joti (2012) Functional Dynamics of Proteins. Computational and Mathemaical Methods in Medicine, 2012, 1-3
New article published (Apr 2011)