


|
|
Home |
DynOmics 1.0 |
Tutorials |
Theory |
References |
iGNM 2.0 |
Mol. Sizer&Timer |
ANM 2.0 |
Comp. Biol. Lab |
PITT site
|
|
Mobility ( → increase)
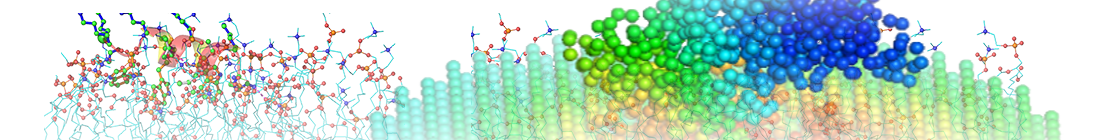
The structure is colored based on the size of fluctuations driven by the slowest two modes.
|
Results for your structure 4B9Q.pdb: Structural Dynamics: Functional Sites Based on Dynamics: Input file and parameters:
PDB title: OPEN CONFORMATION OF ATP-BOUND HSP70 HOMOLOG DNAK PDB: 4B9Q, chain: all, model No.: 1 Exp. method: X-RAY DIFFRACTION (2.40 Å) Cutoff for GNM nodes: 7.3 Å Distance scaling factor: 0 Number of system nodes: 527, number of environment nodes: 76 System of environment ENM: 4-530 residues System of environment ENM: (resid 4 to 530) |

|
Reference: Li,H., Chang,YY, Lee,JY, Bahar,I. and Yang,LW. (2017) DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res., 45, W374-W380. Contact: The server is maintained by Dr. Hongchun Li in the Bahar Lab at the Department of Computational & Systems Biology at the University of Pittsburgh, School of Medicine, and sponsored by the NIH awards #5R01GM099738-04 and #5P41GM103712-03 and the funding #104-2113-M-007-019 from MOST to the Yang lab at the National Tsing Hua University, Taiwan. For questions and comments please contact Hongchun Li.
|