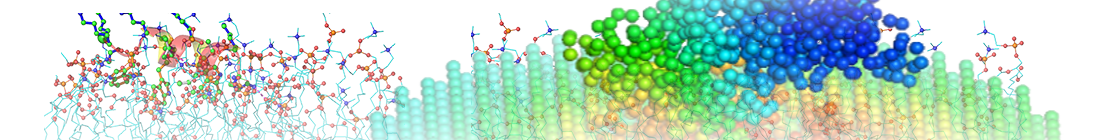



|
| Theoretical B-Factors | Experimental B-Factors |
|
type:
size: px;
Download PDB
|
type:
size: px;
Download PDB
|
|
Mobility ( → increase )
Correlation: 0.71 | |
|
Hide:
Hide/show:
for chain
Export: Click the legends (e.g., Theoretical Chain A) to show/hide the corresponding curves.
|
|
|
Input file and parameters: PDB title: PHOSPHOLIPASE A2 MODIFIED BY PBPB PDB: 1BK9, chain: all, model No.: 1 Exp. method: X-RAY DIFFRACTION (2.00 Å) Number of system nodes: 124 |