Querying the FunSPRINT Server:
1. FunSPRINT predicts the active sites of a given PDB structure or protein sequence. The prediction will be performed for only one structure or sequence. If both the structure and sequence are provided, only the structure will be used for prediction.
2. To provide a PDB structure, user can manually upload a PDB file or enter a PDB ID. Only the manually uploaded structure will be used if both of them are provided.
3. Predictions can be specified for a certain chain, if Chain ID is provided. Otherwise, only the first chain will be used for prediction.
4. To provide a protein sequence, please paste the Fasta format sequence in the textbox. Only one chain is accepted. If structure is provided, sequence will be ignored.
5. Conservation score file: The conservation score file from the Consurf Server (http://consurf.tau.ac.il/) is used in FunSPRINT. Uploading the conservation score file could accelerate the calculation. The file can be downloaded in the result page of Consurf Server from the link: "Amino Acid Conservation Scores, Confidence Intervals and Conservation Colors”
Running Time:
If the conservation score file is provided, FunSPRINT can finish the prediction within a few minutes. Otherwise, prediction might take a few minutes to a few hours. It depends on whether a pre-calculated conservation score file of the given structure/sequence existed or not.
Results Page:
1. For the structure based prediction, the result page consists of two parts:
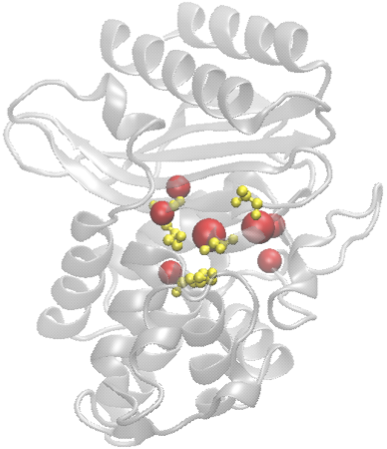
· The structure will be displayed by Jmol (requires Java) and the predicted residues will be labeled and their side-chains will be shown;
· Below it, we will show up to 15 predicted residues ordered by the likelihood that they are active sites. Each residue is associated with a residue ID from the PDB file. Ex. ASP283: Aspartic acid with residue ID 283 in the PDB file.
2. For the sequence based prediction (case I: a enzyme with known active sites matches the sequence): the result page will only list the predicted residue’s position within the query sequence.
3. For the sequence based prediction (case II: none of enzyme with known active sites matches the sequence): FunSPRINT will search against the PDB database to find a similar structure for the sequence then predict the active sites for the searched structure. Sequence alignment among them will be performed. And the residues in the given sequence that align to the predicted active sites in the searched strucutre will be predicted.
· The structure and predicted residues of the searched PDB structure will be reported the same as the structure based prediction;
· Besides, the predicted positions for the query sequence based on the sequence alignment will be listed.
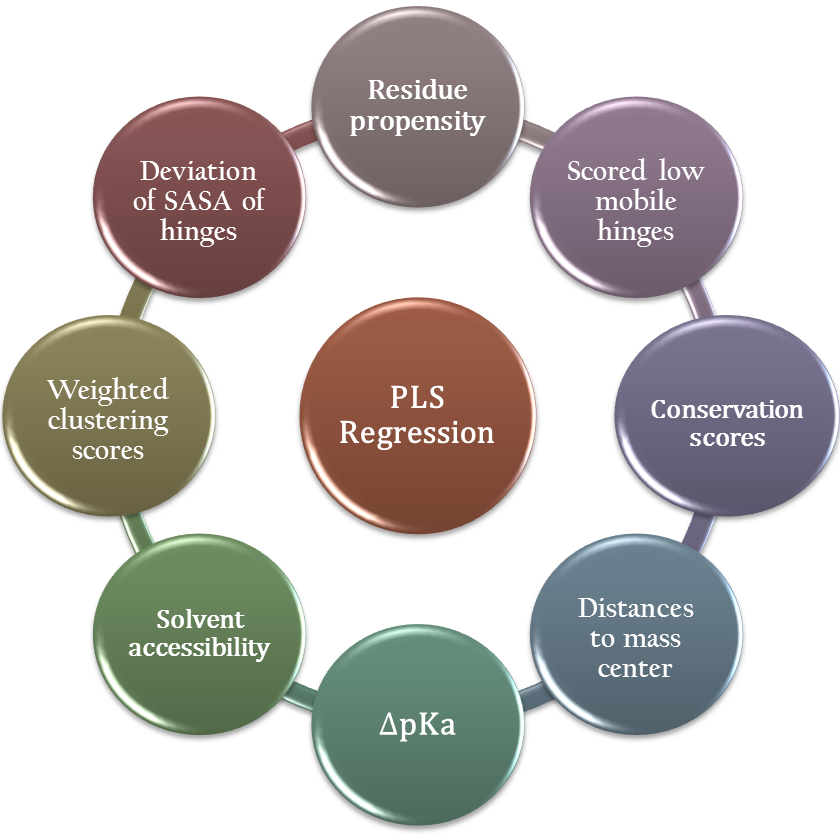
4. Color scheme: The top two of the predicted active sites (hinge centers) are colored in red, 3rd - 7th residues in orange, the 8th onward in green.