



|
|
Home |
DynOmics 1.0 |
Tutorials |
Theory |
References |
iGNM 2.0 |
Mol. Sizer&Timer |
ANM 2.0 |
Comp. Biol. Lab |
PITT site
|
Theory
ANM with membrane (MembrANM)
The ANM represents the complex as a network where each Cα atom serves as a node and uniform network springs connect all the node-pairs within an interaction range, that is, cut-off distance. The overall potential of the system is by definition a summation over all harmonic potentials for the node-pairs (see more details in ANM). MembrANM is an extension of the ANM for modeling the complex (system) and the surrounding lipid bilayer (environment) as a coupled elastic network. See below the structure of 𝛾-secretase with membrane.
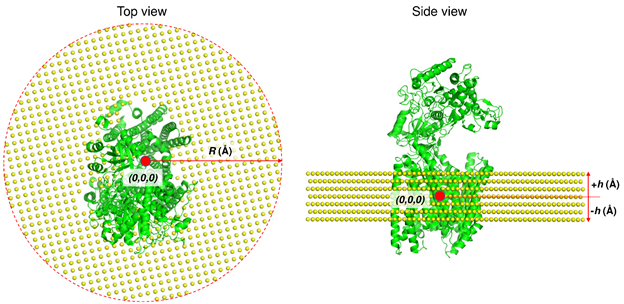
Here, lipid atoms are assumed to occupy a small uniform volume in the membrane and are represented by a single coarse-grained site, which is considered as a node in ANM like Cα atom in protein. The nodes of membrane are constructed using an FCC lattice with the nearest neighbor distance of 6.2 Å. The distance between nodes was selected to give approximately the same node density as in the protein (1). The membrane (in X-Y plane) has a circular shape with R (Å) radius from the center of the protein. The thickness and the position of the membrane (±h Å in Z direction) were obtained from Orientations of Proteins in Membrane (OPM) database (2). membrANM has a network including nodes of membrane as well as nodes of protein, and all the nodes within an interaction range have harmonic interactions.
Using membrANM, we can analyze motions which are more realistic. Including membrane is important in two aspects as follows: 1. Motion between membrane region and extracellular (or intracellular) region: Membrane region is restricted by the membrane but extracellular (or intracellular) region is not, inducing change of their relative motions. 2. Motion within membrane region: The lateral motions of membrane region are restricted by the membrane but its vertical motions are not affected much. Also, membrane region collaborates with the membrane. In 𝛾-secretase case, the top view shape of membrane region is not circle and it looks like horse shoe. The lipids nodes occupy hollow space of the horse shoe, inducing more changes in motion. We show the mode-mode correlations of 𝛾-secretase with and without membrane below.
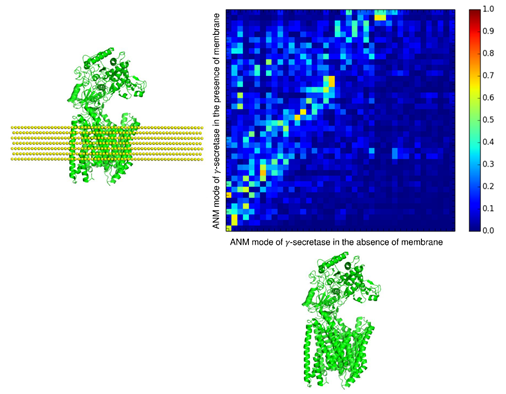
Correlated pairs between the two cases are distributed much, meaning that membrane affected much the motions of the protein (exactly same ANM motions show straight diagonal line). Also, the correlated pairs leaned toward the case of ANM in the presence of membrane. This means that the case with membrane has more various motions than the case without membrane, that is, there are new motions induced by collaboration between membrane and protein.
We can apply this membrANM to various membrane proteins. This will give us more realistic motions and also new motions which are not detectable in ANM without membrane.
Reference
(1) Lezon TR, Bahar I. Constraints imposed by the membrane selectively guide the alternating access dynamics of the glutamate transporter GltPh. Biophys J. 2012;102(6):1331-1340.
(2) Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623-625.